Hierarchical Data Format version 5 interface. More...
#include <opencv2/hdf/hdf5.hpp>
Public Types | |
| enum | { H5_UNLIMITED = -1 , H5_NONE = -1 , H5_GETDIMS = 100 , H5_GETMAXDIMS = 101 , H5_GETCHUNKDIMS = 102 } |
Public Member Functions | |
| virtual | ~HDF5 () |
| virtual void | atdelete (const String &atlabel)=0 |
| virtual bool | atexists (const String &atlabel) const =0 |
| virtual void | atread (double *value, const String &atlabel)=0 |
| virtual void | atread (int *value, const String &atlabel)=0 |
| virtual void | atread (OutputArray value, const String &atlabel)=0 |
| virtual void | atread (String *value, const String &atlabel)=0 |
| virtual void | atwrite (const double value, const String &atlabel)=0 |
| virtual void | atwrite (const int value, const String &atlabel)=0 |
| virtual void | atwrite (const String &value, const String &atlabel)=0 |
| virtual void | atwrite (InputArray value, const String &atlabel)=0 |
| virtual void | close ()=0 |
| Close and release hdf5 object. | |
| virtual void | dscreate (const int n_dims, const int *sizes, const int type, const String &dslabel) const =0 |
| virtual void | dscreate (const int n_dims, const int *sizes, const int type, const String &dslabel, const int compresslevel) const =0 |
| virtual void | dscreate (const int n_dims, const int *sizes, const int type, const String &dslabel, const int compresslevel, const int *dims_chunks) const =0 |
| Create and allocate storage for n-dimensional dataset, single or multichannel type. | |
| virtual void | dscreate (const int rows, const int cols, const int type, const String &dslabel) const =0 |
| virtual void | dscreate (const int rows, const int cols, const int type, const String &dslabel, const int compresslevel) const =0 |
| virtual void | dscreate (const int rows, const int cols, const int type, const String &dslabel, const int compresslevel, const int *dims_chunks) const =0 |
| Create and allocate storage for two dimensional single or multi channel dataset. | |
| virtual void | dscreate (const int rows, const int cols, const int type, const String &dslabel, const int compresslevel, const vector< int > &dims_chunks) const =0 |
| virtual void | dscreate (const vector< int > &sizes, const int type, const String &dslabel, const int compresslevel=HDF5::H5_NONE, const vector< int > &dims_chunks=vector< int >()) const =0 |
| virtual vector< int > | dsgetsize (const String &dslabel, int dims_flag=HDF5::H5_GETDIMS) const =0 |
| Fetch dataset sizes. | |
| virtual int | dsgettype (const String &dslabel) const =0 |
| Fetch dataset type. | |
| virtual void | dsinsert (InputArray Array, const String &dslabel) const =0 |
| virtual void | dsinsert (InputArray Array, const String &dslabel, const int *dims_offset) const =0 |
| virtual void | dsinsert (InputArray Array, const String &dslabel, const int *dims_offset, const int *dims_counts) const =0 |
| Insert or overwrite a Mat object into specified dataset and auto expand dataset size if unlimited property allows. | |
| virtual void | dsinsert (InputArray Array, const String &dslabel, const vector< int > &dims_offset, const vector< int > &dims_counts=vector< int >()) const =0 |
| virtual void | dsread (OutputArray Array, const String &dslabel) const =0 |
| virtual void | dsread (OutputArray Array, const String &dslabel, const int *dims_offset) const =0 |
| virtual void | dsread (OutputArray Array, const String &dslabel, const int *dims_offset, const int *dims_counts) const =0 |
| Read specific dataset from hdf5 file into Mat object. | |
| virtual void | dsread (OutputArray Array, const String &dslabel, const vector< int > &dims_offset, const vector< int > &dims_counts=vector< int >()) const =0 |
| virtual void | dswrite (InputArray Array, const String &dslabel) const =0 |
| virtual void | dswrite (InputArray Array, const String &dslabel, const int *dims_offset) const =0 |
| virtual void | dswrite (InputArray Array, const String &dslabel, const int *dims_offset, const int *dims_counts) const =0 |
| Write or overwrite a Mat object into specified dataset of hdf5 file. | |
| virtual void | dswrite (InputArray Array, const String &dslabel, const vector< int > &dims_offset, const vector< int > &dims_counts=vector< int >()) const =0 |
| virtual void | grcreate (const String &grlabel)=0 |
| Create a group. | |
| virtual bool | hlexists (const String &label) const =0 |
| Check if label exists or not. | |
| virtual void | kpcreate (const int size, const String &kplabel, const int compresslevel=H5_NONE, const int chunks=H5_NONE) const =0 |
| Create and allocate special storage for cv::KeyPoint dataset. | |
| virtual int | kpgetsize (const String &kplabel, int dims_flag=HDF5::H5_GETDIMS) const =0 |
| Fetch keypoint dataset size. | |
| virtual void | kpinsert (const vector< KeyPoint > keypoints, const String &kplabel, const int offset=H5_NONE, const int counts=H5_NONE) const =0 |
| Insert or overwrite list of KeyPoint into specified dataset and autoexpand dataset size if unlimited property allows. | |
| virtual void | kpread (vector< KeyPoint > &keypoints, const String &kplabel, const int offset=H5_NONE, const int counts=H5_NONE) const =0 |
| Read specific keypoint dataset from hdf5 file into vector<KeyPoint> object. | |
| virtual void | kpwrite (const vector< KeyPoint > keypoints, const String &kplabel, const int offset=H5_NONE, const int counts=H5_NONE) const =0 |
| Write or overwrite list of KeyPoint into specified dataset of hdf5 file. | |
Detailed Description
Hierarchical Data Format version 5 interface.
Notice that this module is compiled only when hdf5 is correctly installed.
Member Enumeration Documentation
◆ anonymous enum
| anonymous enum |
| Enumerator | |
|---|---|
| H5_UNLIMITED | The dimension size is unlimited,.
|
| H5_NONE | No compression,.
|
| H5_GETDIMS | Get the dimension information of a dataset.
|
| H5_GETMAXDIMS | Get the maximum dimension information of a dataset.
|
| H5_GETCHUNKDIMS | Get the chunk sizes of a dataset.
|
Constructor & Destructor Documentation
◆ ~HDF5()
|
inlinevirtual |
Member Function Documentation
◆ atdelete()
|
pure virtual |
Delete an attribute from the root group.
- Parameters
-
atlabel the attribute to be deleted.
- Note
- CV_Error() is called if the given attribute does not exist. Use atexists() to check whether it exists or not beforehand.
◆ atexists()
|
pure virtual |
◆ atread() [1/4]
|
pure virtual |
This is an overloaded member function, provided for convenience. It differs from the above function only in what argument(s) it accepts.
◆ atread() [2/4]
|
pure virtual |
Read an attribute from the root group.
- Parameters
-
value address where the attribute is read into atlabel attribute name
The following example demonstrates how to read an attribute of type cv::String:
- Note
- The attribute MUST exist, otherwise CV_Error() is called. Use atexists() to check if it exists beforehand.
◆ atread() [3/4]
|
pure virtual |
Read an attribute from the root group.
- Parameters
-
value attribute value. Currently, only n-d continuous multi-channel arrays are supported. atlabel attribute name.
- Note
- The attribute MUST exist, otherwise CV_Error() is called. Use atexists() to check if it exists beforehand.
◆ atread() [4/4]
This is an overloaded member function, provided for convenience. It differs from the above function only in what argument(s) it accepts.
◆ atwrite() [1/4]
|
pure virtual |
This is an overloaded member function, provided for convenience. It differs from the above function only in what argument(s) it accepts.
◆ atwrite() [2/4]
|
pure virtual |
Write an attribute inside the root group.
- Parameters
-
value attribute value. atlabel attribute name.
The following example demonstrates how to write an attribute of type cv::String:
- Note
- CV_Error() is called if the given attribute already exists. Use atexists() to check whether it exists or not beforehand. And use atdelete() to delete it if it already exists.
◆ atwrite() [3/4]
This is an overloaded member function, provided for convenience. It differs from the above function only in what argument(s) it accepts.
◆ atwrite() [4/4]
|
pure virtual |
Write an attribute into the root group.
- Parameters
-
value attribute value. Currently, only n-d continuous multi-channel arrays are supported. atlabel attribute name.
- Note
- CV_Error() is called if the given attribute already exists. Use atexists() to check whether it exists or not beforehand. And use atdelete() to delete it if it already exists.
◆ close()
|
pure virtual |
Close and release hdf5 object.
◆ dscreate() [1/8]
|
pure virtual |
◆ dscreate() [2/8]
|
pure virtual |
◆ dscreate() [3/8]
|
pure virtual |
Create and allocate storage for n-dimensional dataset, single or multichannel type.
- Parameters
-
n_dims declare number of dimensions sizes array containing sizes for each dimensions type type to be used, e.g., CV_8UC3, CV_32FC1, etc. dslabel specify the hdf5 dataset label. Existing dataset label will cause an error. compresslevel specify the compression level 0-9 to be used, H5_NONE is the default value and means no compression. The value 0 also means no compression. A value 9 indicating the best compression ration. Note that a higher compression level indicates a higher computational cost. It relies on GNU gzip for compression. dims_chunks each array member specifies chunking sizes to be used for block I/O, by default NULL means none at all.
- Note
- If the dataset already exists, an exception will be thrown. Existence of the dataset can be checked using hlexists().
- See example below that creates a 6 dimensional storage space: // open / autocreate hdf5 file// create space for 6 dimensional CV_64FC2 matrixif ( ! h5io->hlexists( "nddata" ) )int n_dims = 5;int dsdims[n_dims] = { 100, 100, 20, 10, 5, 5 };h5io->dscreate( n_dims, sizes, CV_64FC2, "nddata" );elseprintf("DS already created, skipping\n" );// releaseh5io->close();
- Note
- Activating compression requires internal chunking. Chunking can significantly improve access speed both at read and write time, especially for windowed access logic that shifts offset inside dataset. If no custom chunking is specified, the default one will be invoked by the size of whole dataset as single big chunk of data.
- See example of level 0 compression (shallow) using chunking against the first dimension, thus storage will consists of 100 chunks of data: // open / autocreate hdf5 file// create space for 6 dimensional CV_64FC2 matrixif ( ! h5io->hlexists( "nddata" ) )int n_dims = 5;int dsdims[n_dims] = { 100, 100, 20, 10, 5, 5 };int chunks[n_dims] = { 1, 100, 20, 10, 5, 5 };h5io->dscreate( n_dims, dsdims, CV_64FC2, "nddata", 0, chunks );elseprintf("DS already created, skipping\n" );// releaseh5io->close();
- Note
- A value of H5_UNLIMITED inside the sizes array means unlimited data on that dimension, thus it is possible to expand anytime such dataset on those unlimited directions. Presence of H5_UNLIMITED on any dimension requires** to define custom chunking. No default chunking will be defined in unlimited scenario since the default size on that dimension will be zero, and will grow once dataset is written. Writing into dataset that has H5_UNLIMITED on some of its dimension requires dsinsert() instead of dswrite() that allows growth on unlimited dimension instead of dswrite() that allows to write only in predefined data space.
- Example below shows a 3 dimensional dataset using no compression with all unlimited sizes and one unit chunking: // open / autocreate hdf5 fileint n_dims = 3;int chunks[n_dims] = { 1, 1, 1 };int dsdims[n_dims] = { cv::hdf::HDF5::H5_UNLIMITED, cv::hdf::HDF5::H5_UNLIMITED, cv::hdf::HDF5::H5_UNLIMITED };// releaseh5io->close();
◆ dscreate() [4/8]
|
pure virtual |
This is an overloaded member function, provided for convenience. It differs from the above function only in what argument(s) it accepts.
◆ dscreate() [5/8]
|
pure virtual |
This is an overloaded member function, provided for convenience. It differs from the above function only in what argument(s) it accepts.
◆ dscreate() [6/8]
|
pure virtual |
Create and allocate storage for two dimensional single or multi channel dataset.
- Parameters
-
rows declare amount of rows cols declare amount of columns type type to be used, e.g, CV_8UC3, CV_32FC1 and etc. dslabel specify the hdf5 dataset label. Existing dataset label will cause an error. compresslevel specify the compression level 0-9 to be used, H5_NONE is the default value and means no compression. The value 0 also means no compression. A value 9 indicating the best compression ration. Note that a higher compression level indicates a higher computational cost. It relies on GNU gzip for compression. dims_chunks each array member specifies the chunking size to be used for block I/O, by default NULL means none at all.
- Note
- If the dataset already exists, an exception will be thrown (CV_Error() is called).
- Existence of the dataset can be checked using hlexists(), see in this example: // open / autocreate hdf5 file// create space for 100x50 CV_64FC2 matrixif ( ! h5io->hlexists( "hilbert" ) )h5io->dscreate( 100, 50, CV_64FC2, "hilbert" );elseprintf("DS already created, skipping\n" );// releaseh5io->close();
- Note
- Activating compression requires internal chunking. Chunking can significantly improve access speed both at read and write time, especially for windowed access logic that shifts offset inside dataset. If no custom chunking is specified, the default one will be invoked by the size of the whole dataset as a single big chunk of data.
- See example of level 9 compression using internal default chunking: // open / autocreate hdf5 file// create level 9 compressed space for CV_64FC2 matrixif ( ! h5io->hlexists( "hilbert", 9 ) )h5io->dscreate( 100, 50, CV_64FC2, "hilbert", 9 );elseprintf("DS already created, skipping\n" );// releaseh5io->close();
- Note
- A value of H5_UNLIMITED for rows or cols or both means unlimited data on the specified dimension, thus, it is possible to expand anytime such a dataset on row, col or on both directions. Presence of H5_UNLIMITED on any dimension requires to define custom chunking. No default chunking will be defined in the unlimited scenario since default size on that dimension will be zero, and will grow once dataset is written. Writing into a dataset that has H5_UNLIMITED on some of its dimensions requires dsinsert() that allows growth on unlimited dimensions, instead of dswrite() that allows to write only in predefined data space.
- Example below shows no compression but unlimited dimension on cols using 100x100 internal chunking: // open / autocreate hdf5 file// create level 9 compressed space for CV_64FC2 matrixint chunks[2] = { 100, 100 };h5io->dscreate( 100, cv::hdf::HDF5::H5_UNLIMITED, CV_64FC2, "hilbert", cv::hdf::HDF5::H5_NONE, chunks );// releaseh5io->close();
- Note
- It is not thread safe, it must be called only once at dataset creation, otherwise an exception will occur. Multiple datasets inside a single hdf5 file are allowed.
◆ dscreate() [7/8]
|
pure virtual |
This is an overloaded member function, provided for convenience. It differs from the above function only in what argument(s) it accepts.
◆ dscreate() [8/8]
|
pure virtual |
◆ dsgetsize()
|
pure virtual |
Fetch dataset sizes.
- Parameters
-
dslabel specify the hdf5 dataset label to be measured. dims_flag will fetch dataset dimensions on H5_GETDIMS, dataset maximum dimensions on H5_GETMAXDIMS, and chunk sizes on H5_GETCHUNKDIMS.
Returns vector object containing sizes of dataset on each dimensions.
- Note
- Resulting vector size will match the amount of dataset dimensions. By default H5_GETDIMS will return actual dataset dimensions. Using H5_GETMAXDIM flag will get maximum allowed dimension which normally match actual dataset dimension but can hold H5_UNLIMITED value if dataset was prepared in unlimited mode on some of its dimension. It can be useful to check existing dataset dimensions before overwrite it as whole or subset. Trying to write with oversized source data into dataset target will thrown exception. The H5_GETCHUNKDIMS will return the dimension of chunk if dataset was created with chunking options otherwise returned vector size will be zero.
◆ dsgettype()
|
pure virtual |
Fetch dataset type.
- Parameters
-
dslabel specify the hdf5 dataset label to be checked.
Returns the stored matrix type. This is an identifier compatible with the CvMat type system, like e.g. CV_16SC5 (16-bit signed 5-channel array), and so on.
- Note
- Result can be parsed with CV_MAT_CN() to obtain amount of channels and CV_MAT_DEPTH() to obtain native cvdata type. It is thread safe.
◆ dsinsert() [1/4]
|
pure virtual |
◆ dsinsert() [2/4]
|
pure virtual |
◆ dsinsert() [3/4]
|
pure virtual |
Insert or overwrite a Mat object into specified dataset and auto expand dataset size if unlimited property allows.
- Parameters
-
Array specify Mat data array to be written. dslabel specify the target hdf5 dataset label. dims_offset each array member specify the offset location over dataset's each dimensions from where InputArray will be (over)written into dataset. dims_counts each array member specify the amount of data over dataset's each dimensions from InputArray that will be written into dataset.
Writes Mat object into targeted dataset and autoexpand dataset dimension if allowed.
- Note
- Unlike dswrite(), datasets are not created automatically. Only Mat is supported and it must be continuous. If dsinsert() happens over outer regions of dataset dimensions and on that dimension of dataset is in unlimited mode then dataset is expanded, otherwise exception is thrown. To create datasets with unlimited property on specific or more dimensions see dscreate() and the optional H5_UNLIMITED flag at creation time. It is not thread safe over same dataset but multiple datasets can be merged inside a single hdf5 file.
- Example below creates unlimited rows x 100 cols and expands rows 5 times with dsinsert() using single 100x100 CV_64FC2 over the dataset. Final size will have 5x100 rows and 100 cols, reflecting H matrix five times over row's span. Chunks size is 100x100 just optimized against the H matrix size having compression disabled. If routine is called multiple times dataset will be just overwritten: // dual channel hilbert matrixfor(int i = 0; i < H.rows; i++)for(int j = 0; j < H.cols; j++){H.at<cv::Vec2d>(i,j)[0] = 1./(i+j+1);H.at<cv::Vec2d>(i,j)[1] = -1./(i+j+1);count++;}// open / autocreate hdf5 file// optimise dataset by chunksint chunks[2] = { 100, 100 };// create Unlimited x 100 CV_64FC2 spaceh5io->dscreate( cv::hdf::HDF5::H5_UNLIMITED, 100, CV_64FC2, "hilbert", cv::hdf::HDF5::H5_NONE, chunks );// write into first halfint offset[2] = { 0, 0 };for ( int t = 0; t < 5; t++ ){offset[0] += 100 * t;h5io->dsinsert( H, "hilbert", offset );}// releaseh5io->close();
◆ dsinsert() [4/4]
|
pure virtual |
◆ dsread() [1/4]
|
pure virtual |
◆ dsread() [2/4]
|
pure virtual |
◆ dsread() [3/4]
|
pure virtual |
Read specific dataset from hdf5 file into Mat object.
- Parameters
-
Array Mat container where data reads will be returned. dslabel specify the source hdf5 dataset label. dims_offset each array member specify the offset location over each dimensions from where dataset starts to read into OutputArray. dims_counts each array member specify the amount over dataset's each dimensions of dataset to read into OutputArray.
Reads out Mat object reflecting the stored dataset.
- Note
- If hdf5 file does not exist an exception will be thrown. Use hlexists() to check dataset presence. It is thread safe.
- Example below reads a dataset: // open hdf5 file// blank Mat containercv::Mat H;// read hibert dataseth5io->read( H, "hilbert" );// releaseh5io->close();
- Example below perform read of 3x5 submatrix from second row and third element. // open hdf5 file// blank Mat containercv::Mat H;int offset[2] = { 1, 2 };int counts[2] = { 3, 5 };// read hibert dataseth5io->read( H, "hilbert", offset, counts );// releaseh5io->close();
◆ dsread() [4/4]
|
pure virtual |
◆ dswrite() [1/4]
|
pure virtual |
◆ dswrite() [2/4]
|
pure virtual |
◆ dswrite() [3/4]
|
pure virtual |
Write or overwrite a Mat object into specified dataset of hdf5 file.
- Parameters
-
Array specify Mat data array to be written. dslabel specify the target hdf5 dataset label. dims_offset each array member specify the offset location over dataset's each dimensions from where InputArray will be (over)written into dataset. dims_counts each array member specifies the amount of data over dataset's each dimensions from InputArray that will be written into dataset.
Writes Mat object into targeted dataset.
- Note
- If dataset is not created and does not exist it will be created automatically. Only Mat is supported and it must be continuous. It is thread safe but it is recommended that writes to happen over separate non-overlapping regions. Multiple datasets can be written inside a single hdf5 file.
- Example below writes a 100x100 CV_64FC2 matrix into a dataset. No dataset pre-creation required. If routine is called multiple times dataset will be just overwritten: // dual channel hilbert matrixfor(int i = 0; i < H.rows; i++)for(int j = 0; j < H.cols; j++){H.at<cv::Vec2d>(i,j)[0] = 1./(i+j+1);H.at<cv::Vec2d>(i,j)[1] = -1./(i+j+1);count++;}// open / autocreate hdf5 file// write / overwrite dataseth5io->dswrite( H, "hilbert" );// releaseh5io->close();
- Example below writes a smaller 50x100 matrix into 100x100 compressed space optimised by two 50x100 chunks. Matrix is written twice into first half (0->50) and second half (50->100) of data space using offset. // dual channel hilbert matrixfor(int i = 0; i < H.rows; i++)for(int j = 0; j < H.cols; j++){H.at<cv::Vec2d>(i,j)[0] = 1./(i+j+1);H.at<cv::Vec2d>(i,j)[1] = -1./(i+j+1);count++;}// open / autocreate hdf5 file// optimise dataset by two chunksint chunks[2] = { 50, 100 };// create 100x100 CV_64FC2 compressed spaceh5io->dscreate( 100, 100, CV_64FC2, "hilbert", 9, chunks );// write into first halfint offset1[2] = { 0, 0 };h5io->dswrite( H, "hilbert", offset1 );// write into second halfint offset2[2] = { 50, 0 };h5io->dswrite( H, "hilbert", offset2 );// releaseh5io->close();
◆ dswrite() [4/4]
|
pure virtual |
◆ grcreate()
|
pure virtual |
Create a group.
- Parameters
-
grlabel specify the hdf5 group label.
Create a hdf5 group with default properties. The group is closed automatically after creation.
- Note
- Groups are useful for better organising multiple datasets. It is possible to create subgroups within any group. Existence of a particular group can be checked using hlexists(). In case of subgroups, a label would be e.g: 'Group1/SubGroup1' where SubGroup1 is within the root group Group1. Before creating a subgroup, its parent group MUST be created.
In this example, Group1 will have one subgroup called SubGroup1:
// "/" means the root group, which is always presentif (!h5io->hlexists("/Group1"))h5io->grcreate("/Group1");elsestd::cout << "/Group1 has already been created, skip it.\n";// Note that Group1 has been created above, otherwise exception will occurif (!h5io->hlexists("/Group1/SubGroup1"))h5io->grcreate("/Group1/SubGroup1");elsestd::cout << "/Group1/SubGroup1 has already been created, skip it.\n";h5io->close();The corresponding result visualized using the HDFView tool is
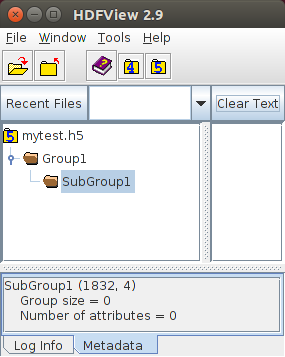
- Note
- When a dataset is created with dscreate() or kpcreate(), it can be created within a group by specifying the full path within the label. In our example, it would be: 'Group1/SubGroup1/MyDataSet'. It is not thread safe.
◆ hlexists()
|
pure virtual |
Check if label exists or not.
- Parameters
-
label specify the hdf5 dataset label.
Returns true if dataset exists, and false otherwise.
- Note
- Checks if dataset, group or other object type (hdf5 link) exists under the label name. It is thread safe.
◆ kpcreate()
|
pure virtual |
Create and allocate special storage for cv::KeyPoint dataset.
- Parameters
-
size declare fixed number of KeyPoints kplabel specify the hdf5 dataset label, any existing dataset with the same label will be overwritten. compresslevel specify the compression level 0-9 to be used, H5_NONE is default and means no compression. chunks each array member specifies chunking sizes to be used for block I/O, H5_NONE is default and means no compression.
- Note
- If the dataset already exists an exception will be thrown. Existence of the dataset can be checked using hlexists().
- See example below that creates space for 100 keypoints in the dataset: // open hdf5 fileif ( ! h5io->hlexists( "keypoints" ) )h5io->kpcreate( 100, "keypoints" );elseprintf("DS already created, skipping\n" );
- Note
- A value of H5_UNLIMITED for size means unlimited keypoints, thus is possible to expand anytime such dataset by adding or inserting. Presence of H5_UNLIMITED require to define custom chunking. No default chunking will be defined in unlimited scenario since default size on that dimension will be zero, and will grow once dataset is written. Writing into dataset that have H5_UNLIMITED on some of its dimension requires kpinsert() that allow growth on unlimited dimension instead of kpwrite() that allows to write only in predefined data space.
- See example below that creates unlimited space for keypoints chunking size of 100 but no compression: // open hdf5 fileif ( ! h5io->hlexists( "keypoints" ) )elseprintf("DS already created, skipping\n" );
◆ kpgetsize()
|
pure virtual |
Fetch keypoint dataset size.
- Parameters
-
kplabel specify the hdf5 dataset label to be measured. dims_flag will fetch dataset dimensions on H5_GETDIMS, and dataset maximum dimensions on H5_GETMAXDIMS.
Returns size of keypoints dataset.
- Note
- Resulting size will match the amount of keypoints. By default H5_GETDIMS will return actual dataset dimension. Using H5_GETMAXDIM flag will get maximum allowed dimension which normally match actual dataset dimension but can hold H5_UNLIMITED value if dataset was prepared in unlimited mode. It can be useful to check existing dataset dimension before overwrite it as whole or subset. Trying to write with oversized source data into dataset target will thrown exception. The H5_GETCHUNKDIMS will return the dimension of chunk if dataset was created with chunking options otherwise returned vector size will be zero.
◆ kpinsert()
|
pure virtual |
Insert or overwrite list of KeyPoint into specified dataset and autoexpand dataset size if unlimited property allows.
- Parameters
-
keypoints specify keypoints data list to be written. kplabel specify the target hdf5 dataset label. offset specify the offset location on dataset from where keypoints will be (over)written into dataset. counts specify the amount of keypoints that will be written into dataset.
Writes vector<KeyPoint> object into targeted dataset and autoexpand dataset dimension if allowed.
- Note
- Unlike kpwrite(), datasets are not created automatically. If dsinsert() happen over outer region of dataset and dataset has been created in unlimited mode then dataset is expanded, otherwise exception is thrown. To create datasets with unlimited property see kpcreate() and the optional H5_UNLIMITED flag at creation time. It is not thread safe over same dataset but multiple datasets can be merged inside single hdf5 file.
- Example below creates unlimited space for keypoints storage, and inserts a list of 10 keypoints ten times into that space. Final dataset will have 100 keypoints. Chunks size is 10 just optimized against list of keypoints. If routine is called multiple times dataset will be just overwritten: // generate 10 dummy keypointsstd::vector<cv::KeyPoint> keypoints;for(int i = 0; i < 10; i++)keypoints.push_back( cv::KeyPoint(i, -i, 1, -1, 0, 0, -1) );// open / autocreate hdf5 file// create unlimited size space with chunk size of 10h5io->kpcreate( cv::hdf::HDF5::H5_UNLIMITED, "keypoints", -1, 10 );// insert 10 times same 10 keypointsfor(int i = 0; i < 10; i++)h5io->kpinsert( keypoints, "keypoints", i * 10 );// releaseh5io->close();
◆ kpread()
|
pure virtual |
Read specific keypoint dataset from hdf5 file into vector<KeyPoint> object.
- Parameters
-
keypoints vector<KeyPoint> container where data reads will be returned. kplabel specify the source hdf5 dataset label. offset specify the offset location over dataset from where read starts. counts specify the amount of keypoints from dataset to read.
Reads out vector<KeyPoint> object reflecting the stored dataset.
- Note
- If hdf5 file does not exist an exception will be thrown. Use hlexists() to check dataset presence. It is thread safe.
- Example below reads a dataset containing keypoints starting with second entry: // open hdf5 file// blank KeyPoint containerstd::vector<cv::KeyPoint> keypoints;// read keypoints starting second oneh5io->kpread( keypoints, "keypoints", 1 );// releaseh5io->close();
- Example below perform read of 3 keypoints from second entry. // open hdf5 file// blank KeyPoint containerstd::vector<cv::KeyPoint> keypoints;// read three keypoints starting second oneh5io->kpread( keypoints, "keypoints", 1, 3 );// releaseh5io->close();
◆ kpwrite()
|
pure virtual |
Write or overwrite list of KeyPoint into specified dataset of hdf5 file.
- Parameters
-
keypoints specify keypoints data list to be written. kplabel specify the target hdf5 dataset label. offset specify the offset location on dataset from where keypoints will be (over)written into dataset. counts specify the amount of keypoints that will be written into dataset.
Writes vector<KeyPoint> object into targeted dataset.
- Note
- If dataset is not created and does not exist it will be created automatically. It is thread safe but it is recommended that writes to happen over separate non overlapping regions. Multiple datasets can be written inside single hdf5 file.
- Example below writes a 100 keypoints into a dataset. No dataset precreation required. If routine is called multiple times dataset will be just overwritten: // generate 100 dummy keypointsstd::vector<cv::KeyPoint> keypoints;for(int i = 0; i < 100; i++)keypoints.push_back( cv::KeyPoint(i, -i, 1, -1, 0, 0, -1) );// open / autocreate hdf5 file// write / overwrite dataseth5io->kpwrite( keypoints, "keypoints" );// releaseh5io->close();
- Example below uses smaller set of 50 keypoints and writes into compressed space of 100 keypoints optimised by 10 chunks. Same keypoint set is written three times, first into first half (0->50) and at second half (50->75) then into remaining slots (75->99) of data space using offset and count parameters to settle the window for write access.If routine is called multiple times dataset will be just overwritten: // generate 50 dummy keypointsstd::vector<cv::KeyPoint> keypoints;for(int i = 0; i < 50; i++)keypoints.push_back( cv::KeyPoint(i, -i, 1, -1, 0, 0, -1) );// open / autocreate hdf5 file// create maximum compressed space of size 100 with chunk size 10h5io->kpcreate( 100, "keypoints", 9, 10 );// write into first halfh5io->kpwrite( keypoints, "keypoints", 0 );// write first 25 keypoints into second halfh5io->kpwrite( keypoints, "keypoints", 50, 25 );// write first 25 keypoints into remained space of second halfh5io->kpwrite( keypoints, "keypoints", 75, 25 );// releaseh5io->close();
The documentation for this class was generated from the following file:
- opencv2/hdf/hdf5.hpp
 1.9.6
1.9.6